An RNA Microcode Underlying Embryonic Development
The microcode that bootstraps embryonic development
The discovery of the microcode controlling gene expression during development was quite unexpected and very surprising. Even step we took led to a new and exciting finding.”
CHARLESTOWN, MA, UNITED STATES, March 3, 2023 /EINPresswire.com/ -- A peer-revied paper released today in the International Journal of Molecular Sciences uncovers how a small RNAs based microcode directs embryonic development. The current works suggests that the microcode embedded in small RNAs bootstraps gene expression at early stages of embryonic development. Conceptually, this design is similar to the way a computer uses a bootloader to install a higher level operating system. The findings reported have high statistical significance, exceeding the 5 sigma threshold set for discovery in the physical sciences, something that is not usual for the biological sciences.— Alan Herbert
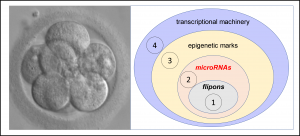
The small RNAs examined in the study are called microRNAs. They target DNA sequences called flipons that can form alternative DNA structures like left-handed Z-DNA and four stranded quadruplexes. By setting the DNA conformation, the RNA microcode changes the read out of information from the genome. The work focused on the microcode that is highly conserved and embedded in the microRNAs of many species. The flipons targeted by those microRNAs are present in the promoters of genes with key roles in specifying embryonic development.
The use of microRNAs (Figure 1) to control the shape of flipons in DNA promoters is a different way to regulate gene expression. The RNA microcode recognize DNA in a sequence-specific manner. The interaction then specifies the 3D structure of the flipon bound. The readout of genetic information then relies on proteins specific for each flipon structure. A good analogy is the way ribosomes works. A ribosome can make many different proteins, but will only extend a peptide if the correct 3D-structure is formed by a codon, anticodon pair. The codon triplet sequence does not matter, nor does the amino acid attached to the tRNA. Only the structure formed by the codon and anticodon is important. In a similar manner, proteins will require that flipons adopt a particular conformation before they will readout a particular set of information from the genome.
The mechanism based on microRNA and DNA structure contrasts with current models that focus on the sequence-specific recognition of DNA by proteins to control gene expression. A major difference between the two strategies is how quickly each evolves. An RNA microcode can change faster than is possible with sequence-specific binding proteins. Elaborating RNAs to target new regions in the genome is a lot less complex than making and identifying protein mutations that produce the same outcome. The problem with the protein focused strategy is that each new mutation runs the risk of interfering with some other function the protein performs. With the RNA microcode strategy, many different microRNAs can be evaluated in parallel. Those producing successful adaptations will be favored by natural selection without the need to discard the microRNAs that were useful in the past. Over time, built-for-purpose proteins can take over the role of microRNAs to optimize outcomes.
The work performed by the team involved mapping conserved microRNAs to flipons. The focus was on the microRNAs that are highly conserved in placental animals. These microRNAs bind to DNA at sites called flipons, sequences that can change from one conformation to another within a cell. Flipons can form alternative structures like the left-handed Z-DNA or the four stranded G4 quadruplex fold. These alternative conformations are of higher energy that the right-handed Watson and Crick B-DNA. Normally only active genes produce sufficient energy to power the flip. The structure formed flag these regions of the genomes for recruitment of specific proteins to these locations. The conserved miR provide a mechanism to direct this process. Many microRNAs are transmitted from the parents to embryos via sperm and eggs and are thought to influence the early stages of development. The parental microRNAs then have the potential to impact offspring phenotype.
InsideOutBio is an early-stage biotech company developing therapeutics for the treatment of cancer The access to the enormous databases created by collaborative international efforts has helped the InsideOutBio scientists make fundamental discoveries such as those reported by Dr. Herbert, Mr. Fed0rov and Dr. Poptsova in the paper. Previously Dr. Herbert discovered a family of proteins that bind the left-handed Z-DNA conformation and performed pioneering human genetic studies in the Framingham Heart Study. InsideOutBio is privately funded.
Fedor Pavlov, Dimitrii Konovalov and Maria Poptsova are supported by the Basic Research Program of the National Research University Higher School of Economics, for which Dr. Herbert is an International Supervisor.
Alan Herbert
InsideOutBio, Inc
+16178007531 ext.
email us here
Legal Disclaimer:
EIN Presswire provides this news content "as is" without warranty of any kind. We do not accept any responsibility or liability for the accuracy, content, images, videos, licenses, completeness, legality, or reliability of the information contained in this article. If you have any complaints or copyright issues related to this article, kindly contact the author above.